Speeding up summary.survfit
I’m working on a simulation study to benchmark a score test for centre effects in competing risk models. The particular model we’re using (Fine and Gray) relies on weights calculated from the Kaplan-Meier estimate of the censoring distribution, and thus makes repeated calls to summary.survfit.
Although a single call to summary.survfit does not take long, the calls can add up. And in my case they do. My supervisor ran a partial simulation study a few years back, and the code took 47 hours to run.
In the interest of graduating this semester, I set out to find ways of speeding up the process. The latest version of RStudio includes support for memory profiling with profvis, making it easy to find bottlenecks in your R code.
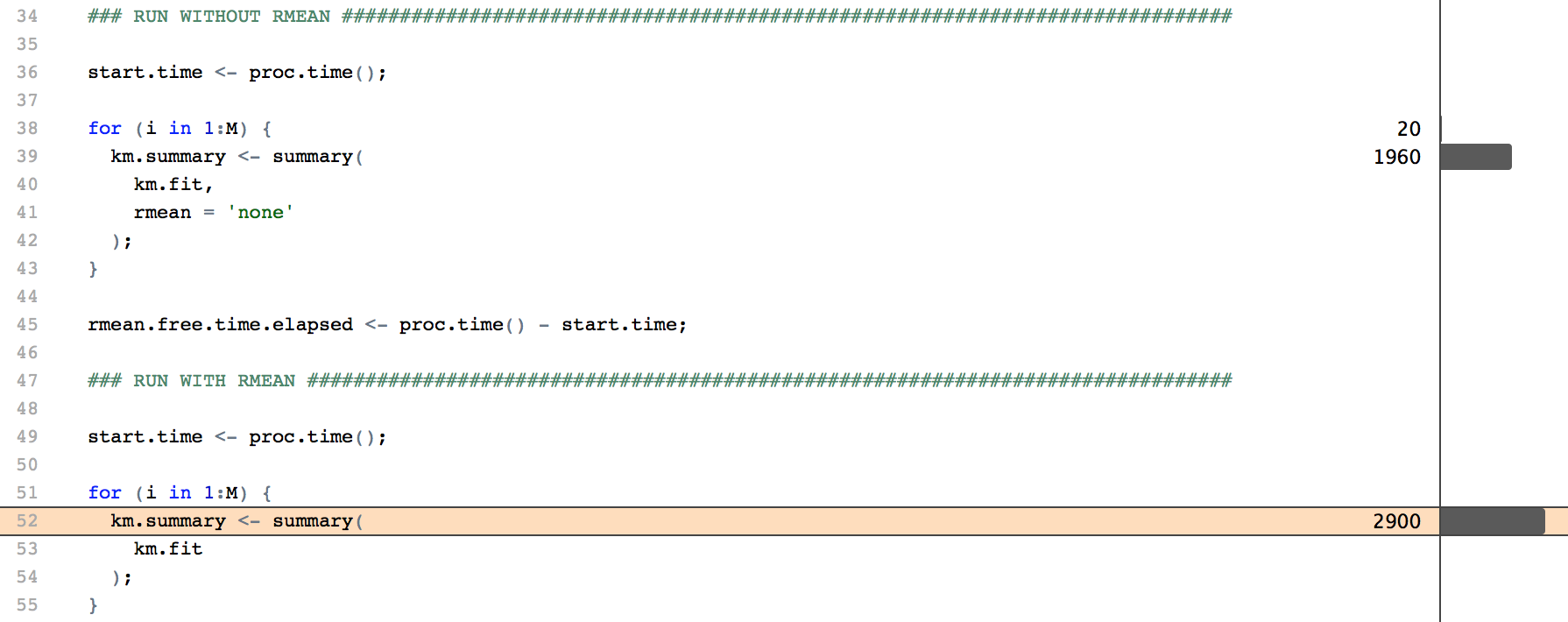
After profiling the code, I found a surprisingly efficient way to speed up the process. By default, summary.survfit calculates the restricted mean of the survival times (RMST). This is the expected survival within a given time frame, for example the 5-year life expectancy.
While RMST is an interesting concept in its own right, it is entirely irrelevant to my competing risk study. The calculation can be turned off by passing rmean = 'none' in the summary call, or setting the option survfit.rmean = 'none'.
In my case, this resulted in a 40% reduction in the time required for a single function call, and a 9% reduction for my full script. Realistically, this won’t make much of a difference in most cases. But after spending most of my afternoon on the investigation, it was a very satisfying result!